30 years later: the dynamical transition studied under high hydrostatic pressure
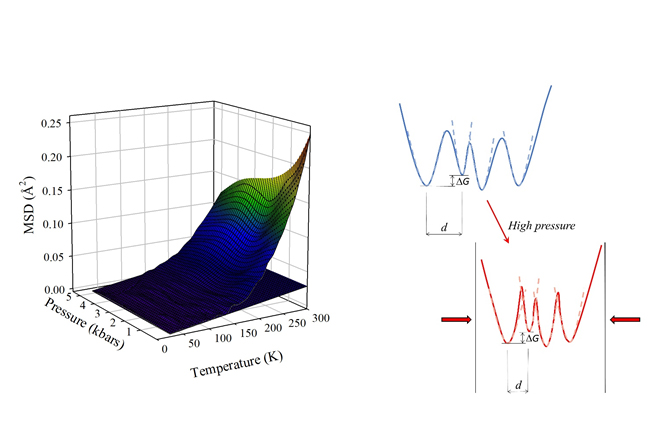
Mean Square Displacements (MSD) obtained from the analysis in terms of a two-state model and schematic representation of the effects of pressure on the protein energy landscape
Protein dynamics are characterized by fluctuations among different conformational substates, i.e. the different minima of their energy landscape. At temperatures above ~200K, these fluctuations lead to a steep increase in the thermal dependence of all dynamical properties, a phenomenon known as the Protein Dynamical Transition. In spite of many studies, little is known about the effects of pressure on these processes, investigated mostly near room temperature. Neutron scattering is an ideal technique and elastic Incoherent Neutron Scattering, in particular, probes directly the Mean Square Displacements (MSD) of protein hydrogen atoms. Since hydrogen atoms make up about half the atoms in a protein and are almost uniformly distributed within the molecule, this technique gives a global view of protein internal motions.
The effects of pressure on the energy landscape of proteins and on their internal dynamical properties are the subject of a study recently published in Scientific Reports. The study arises from a collaboration between researchers from ILL and Université Grenoble Alpes, from the Institute of Biophysics of the Italian National Research Council (IBF-CNR, Palermo, Italy), and from the Physics and Chemistry Department of the University of Palermo (Italy). In this work, the dynamics of myoglobin was studied in a wide range of temperature and pressure. For the experiments, performed on the IN13 backscattering spectrometer, a highly viscous, deuterated, glycerol-protein-water sample was used, suitable for pressure transmission at cryogenic temperatures and for a precise detection of the relevant protein internal motions. The results show that high pressure reduces protein motions, but does not affect the onset temperature for the Protein Dynamical Transition. This indicates that the energy differences and barriers between conformational substates do not change with pressure. Instead, the observed reduction of protein internal motions observed at high pressure must be ascribed to a corresponding reduction in the average structural differences between the accessible conformational substates, which means an increase of the effective roughness of the free energy landscape of the system (see figure).
Since the dynamical properties of proteins are coupled over decades in time and self-similar, the results obtained in this study will help to build a unique scenario for the effects of pressure on protein dynamics. A more rugged energy landscape can clearly have a strong influence on the sub-nanosecond dynamics of proteins, as in the case of this study, but it may also affect motions occurring on much longer time scales, up to those required for protein function or even for processes like folding and unfolding.
This study is published in Scientific Reports.

Fig 1: Mean Square Deplacements (MSD) obtained from the analysis in terms of a two-state model and schematic representation of the effects of pressure on the protein energy landscape
Contact:Judith Peters (UGA, LiPhy and ILL)
