New technique reveals how bacteria initiate infection in unprecedented detail
According to the World Health Organisation, antimicrobial resistance is one of the top 10 threats facing humanity, as a significant danger to both global health and food security. It is rising to concerning levels across the world.
Understanding the interaction between bacteria and human cells at the point of infection is vital for finding solutions to antimicrobial resistance. This relationship is key for the design of novel drugs that can help find ways to reduce the impact of resistant strains in the body, by stopping infections from occurring in the first place.
Researchers at Institut Laue Langevin (ILL) and Centre de Recherches sur les Macromolécules Végétales (CERMAV) have used neutrons to unpick the binding process that instigates infection in host cells. They observed lectins, which are responsible for facilitating this process across a range of microorganisms, including viruses as well as bacteria.
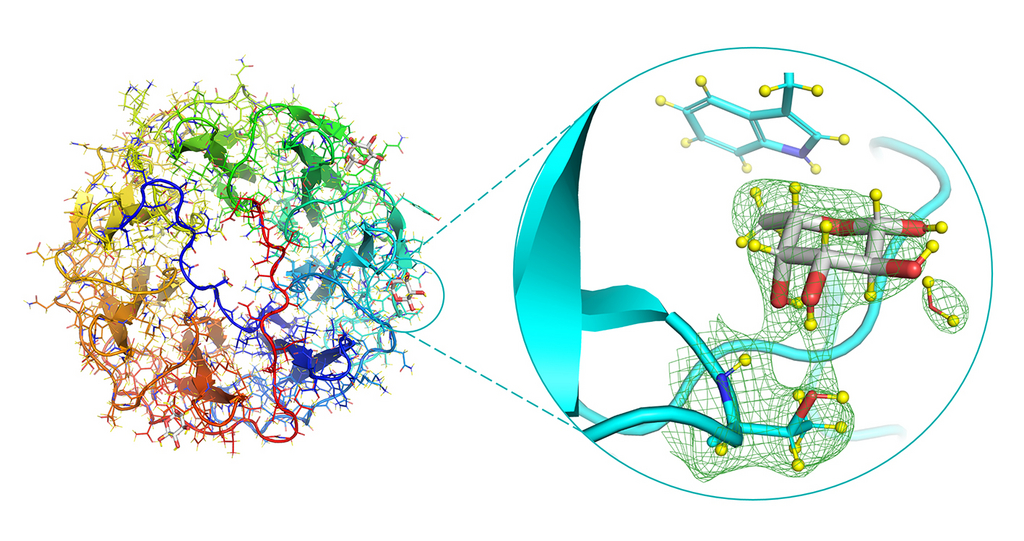
Neutron structure of the PLL/fucose complex, focusing on the fucose-binding site (right). The green mesh represents the neutron density.
Lectins bind to sugars (glycans) on the surface of the host cell and dictate the features that can make these tiny organisms so deadly – how infectious or adhesive they can be, and even how they hinder the actions of other microbes. This process is facilitated by the method that lectins use to expertly recognise the sugars on the surface of the cell, even though these can be extremely varied. They then start to take control of the tissues. Without this ability to decipher what the sugar is, research has shown that bacteria significantly decreases in its potency of infection.
In order to better understand this process, the scientists used neutron macromolecular crystallography to reveal the full atomic structure of a bacterial lectin interacting with fucose (a simple sugar that is one of the building blocks of more complex sugars). Neutrons are an extremely effective tool for observing hydrogen bonds, and therefore are perfect for studying protein-carbohydrate (protein-sugar) interactions. The atomic make-up means that the neutrons scatter strongly from the hydrogen’s nucleus, which can be used to create a map of the bonds involved in the interactions. It is this full atomic detail which will help improve our understanding for structure-based drug design.
The researchers were also able to see the interactions in unprecedented detail due to a new method used. Both the lectin and the sugar were perdeuterated in vivo. Using bacteria that have been engineered to produce the sugar of interest, fucose, the hydrogen atoms were replaced with deuterium atoms to allow for greater contrast and therefore enhanced detail in the experiment. This approach to preparing the experiment is not yet commonly done, but could be invaluable in increasing the visibility of the binding processes and enabling effective modelling of the bacteria and sugar interaction in other models for future drug development.
In addition to improved models of protein-sugar interactions, this research will help scientists to find alternative approaches that will prevent bacteria from binding to human cells, also known as antiadhesive therapy. This is a particularly promising route as it does not itself require antibiotics to prevent infections, and therefore does not further promote antimicrobial resistance.
“There are far more pathogenic bacteria we could study in order to better understand this relationship. Sugar binding proteins are an ideal target for neutron crystallography, as many interactions with the sugar involve hydrogen atoms. Here we have been able to describe a complete hydrogen-bonding network between the bacterial lectin and fucose in unprecedented detail.” Lukáš Gajdoš, Doctoral Student, Institut Laue-Langevin.
- Production of perdeuterated fucose from glyco-engineered bacteria (2021) : DOI: 10.1093/glycob/cwaa059
- Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions (2021) : DOI: doi.org/10.1016/j.str.2021.03.003
ILL Instrument: LADI-III, which delivers a high flux of neutrons and only requires a sample size of 0.05-0.1 mm3 in order to get results. The samples were prepared using the Deuteration Laboratory (D-Lab) platform in the ILL's Life Sciences group.
Contact:.” Lukáš Gajdoš (ILL)
