Protein dynamics: dancing at different rhythms
Proteins perform a vast range of functions in living organisms. They are known to be dynamical entities, moving and changing shape in ways that are crucial to their functioning. Protein dynamics have been previously investigated at various timescales. Nanosecond and picosecond dynamics have been assigned to local motion, while slower dynamics have been attributed to larger conformational changes. The goal of this study was to complete the mapping of the dynamics of the well-known protein Hsp90 by studying the 5-500 ns timescale using a combination of state-of-the-art complementary techniques, and to take the first steps towards understanding how this timescale relates to the slower dynamics known to drive the protein’s functioning.
What is this all about?
Imagine you are the size of an atom. Floating in front of you there is a protein molecule. Thousands of atoms are bound in certain ways, organised in regular and repetitive patterns that are not easy to fully grasp at first sight. And everything moves: atoms oscillate in pairs or groups; some of them drift, on their own or in groups. Sometimes, some parts seem to bend or vibrate in coherent ways, and at a slower pace the whole thing evolves in space, rotating and wiggling as a whole or in chunks. There are so many parts, types of movement and widely different rhythms that it is hard to make sense of it. Regularities and sequences are hard to riddle. You really need to focus on one timescale at a time, split the movements into types and take notes! It is probably best to record the sequences in different ways and then combine and analyse all the information. Putting it all together would make it possible to build a map of how the protein moves at different timescales to aid an understanding of how the different movement patterns relate to each other and to the way in which the protein functions. Now imagine you are back to your normal size and try to do the same thing. The complexity we just described is still there. Just add to it the fact that you can’t even see the molecule. The ways to record and analyse data will have to be different, relying on other wavelengths and processes – such as neutron scattering and complementary techniques.
The goal of this study was precisely to complete a map of the dynamics of the protein Hsp90 by studying the 100-200 ns timescale by using a combination of state-of-the-art complementary techniques and take the first steps towards understanding how dynamics at this timescale relates to the slower, conformal dynamics known to drive the protein’s functioning. Let’s look at it step by step.
Protein dynamics and timescales

Proteins are large molecules made from long amino acid chains performing a vast range of functions in living organisms. Many proteins fold up into stable 3D shapes defined by their amino acid sequences. However, the conformation may change and this is crucial to the functioning of the protein. A great deal of the complexity in understanding protein function results from the involvement of dynamic processes occurring at a broad range of timescales.
Protein dynamics have been previously investigated at various timescales. Numerous studies have provided important insights into dynamics at the microsecond to second timescale. Nanosecond and picosecond dynamics have been assigned to local motion, while slower dynamics have been attributed to larger conformational changes. But how can fast (local) fluctuations lead to slow (global) changes? While this remains largely unknown, there are strong indications that the dynamics within conformational states, likely at timescales of hundreds of nanoseconds, might enable conformational transitions related to function. If this is the case, this timescale plays a crucial role and deserves further investigation.
This challenge was addressed using a well-known protein: heat shock protein 90 (Hsp90). Heat shock proteins are produced by cells in response to stress conditions (such as heat, cold,UV light or wound healing). Many of them perform molecular chaperone functions, i.e. they assist protein folding and help refold proteins that have been damaged. Hsp90 is a central regulatory element in various cellular processes. Misfolded proteins are often linked to neurodegenerative diseases and cancer, which makes Hsp90 a promising target for research into new drugs. Throughout its chaperoning cycle, Hsp90 transitions between open and closed molecular conformations at timescales from milliseconds to several minutes that have been well investigated. Local dynamics at the picosecond timescale have also been reported before for Hsp90. To complete the picture, the nanosecond timescales still need to be addressed.
A combined approach
The goal was thus to shed light on the previously unaddressed nanosecond dynamics of the molecular chaperone protein Hsp90. The complexity of the challenge called for an integrative approach combining different, often complementary experimental and computer simulation techniques. Besides Hsp90, the basic ingredients were advanced molecular fluorescence spectroscopy, neutron scattering and all-atom molecular dynamics simulations.
Advanced molecular fluorescence spectroscopy
Fluorescence is a type of luminescence caused by photons exciting a molecule. As the excited molecule returns to ground state, it emits a photon of lower energy (longer wavelength) than the absorbed photon. Spectroscopy refers to the analyses of the absorption and emission of radiation, namely of the energy (wavelength) and intensity variations in the process. In fluorescence correlation spectroscopy (FCS), typically a few molecules are observed and the measured fluorescence intensity fluctuations (due to physical or chemical reactions) are analysed using their temporal correlation. The way in which signals with independent causes are superimposed and can be disentangled depends on the difference in their timescales. In this study, the authors took advantage of advanced techniques. Most surprisingly, pronounced dynamics were observed around 150 ns.
Neutron scattering
Neutrons offer unique possibilities in the study of materials’ structure and dynamics. In a typical experiment, neutrons scattered off the sample are measured as a function of the change in their direction (related to momentum transfer) and energy (energy transfer). While the first tells us about the structure of the scattering material, the energy transfer dependence tells us about its dynamics. Depending on the energy and timescales of interest, different scattering methods are available. Due to significantly lower energy as compared to photons with nm wavelength, neutrons are an excellent probe for studying low energy excitations such as molecular rotations and diffusion. The technique proved to be efficient in characterising the motion of individual atoms in proteins. Recent advances in neutron instrumentation allow us to separate the different superimposed contributions.
The state of the art of neutron scattering instruments is constantly evolving, in particular improving the resolution and accelerating the rate at which data can be acquired. Neutron spin-echo and backscattering spectrometers probe the dynamics in materials at the picosecond and nanosecond timescales and provide spatial information on motion at the nanometre scale. Neutron spin-echo experiments were performed at the spin-echo spectrometer IN15 to investigate dynamics at timescales from few to several hundred nanoseconds. Quasi-Elastic Neutron Backscattering data were measured at the backscattering spectrometer IN16B, observing the nanosecond timescale. The samples used in NSE and NBS were identical and measured at the same temperature (295 K) to ensure data consistency. Structural properties have been investigated with small angle neutron scattering on D11.
All-atom molecular dynamics simulations and interpretation of the results
Computer simulations of molecular dynamics (MD) simulations are ideal to predict molecular details at these fast timescales and a precious tool in the interpretation of experimental data. Here, the motions of all atoms within a protein are predicted based on their inter-atom forces with femtosecond resolution. The combination of this theoretical approach with experimental measurements allowed us to distinguish internal dynamics from global rotation, although they occur at similar timescales. The calculated dynamics shows a complex pattern of diffusive modes, which can be described with our integrative approach covering a wide range of time and length scales. Global real-space movements are assigned to dynamic modes at this timescale.
As a first step towards confirming a possible functional relevance of dynamics at the 150 ns timescale, the authors verified that these dynamic modes depend on the conformational and functional state of Hsp90, in very good agreement with simulations. Molecule-spanning dynamics on the 150 ns timescale might be precursors to large conformational changes between two equilibrium states.
Together with the previously published dynamics at slower timescales, the results obtained complete a comprehensive picture of Hsp90’s dynamics from the nanosecond to second timescale. Altogether, data are best described by fast molecule-spanning dynamics, which precede larger conformational changes in Hsp90 and might be the molecular basis for slow global changes.
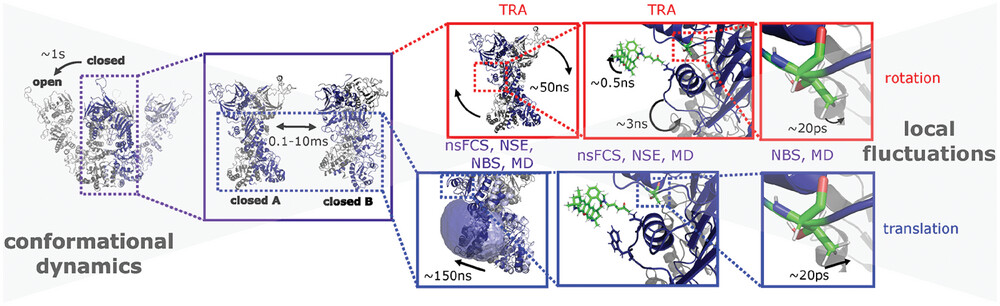
The picture of Hsp90 dynamics has been completed by accessing the nanosecond timescale, which revealed molecule-spanning dynamics on the order of ~150 ns. Disentanglement of rotational and translational dynamics (red and blue frames, respectively) was achieved by combining nanosecond fluorescence correlation spectroscopy (nsFCS), time-resolved anisotropy (TRA), neutron spin-echo spectroscopy (NSE), neutron backscattering spectroscopy (NBS) and molecular dynamics (MD) simulations. The timescales (from experiments) of the respective dynamics are given in the boxes. TRA is only sensitive to rotations, while all other methods are sensitive to rotation and translation.
Reference: Benedikt Sohmen, Christian Beck, Veronika Frank, Tilo Seydel, Ingo Hoffmann, Bianca Hermann, Mark Nüesch, Marco Grimaldo, Frank Schreiber, Steffen Wolf, Felix Roosen-Runge, and Thorsten Hugel, “The Onset of Molecule-Spanning Dynamics in Heat Shock Protein Hsp90”, Adv. Sci. 10 (2023) 2304262.
https://doi.org/10.1002/advs.202304262
ILL Contact: Christian Beck

