


Next: MAGNETIC STRUCTURES AND STRUCTURE FACTORS
Up: LEAST SQUARES REFINEMENT USING
Previous: Making Other Least Squares
Subsections
Structure Factor Least Squares with multiple Data Sources
As of Version 4.4 of CCSL the set of structure factor least squares programs have been reorganised to allow
data from more than one source to be refined concurrently. To facilitate this,
provision has been made to attach source dependent information such as crystal orientation, wavelength, neutron polarisation etc, to the files containing the experimental data. This information is found in lines at the start of the data files begining with a # followed by a word indicating the type of information and the information itself. The words following the # which can currently be recognised are:
- Polarisation followed by 5 numbers: the Up and Down polarising efficiencies
and the polarisation direction in orthogonal crystal coordinates.
- Wavelength followed by its value and, if needed, the absorption
coefficient.
- Orientation followed by 9 components of the matrix giving the directions of the diffractometer axes in orthogonal crystal coordinates.
Only the first 5 characters of the words are matched, their case is unimportant.
The CCSL programs
ABSMSF,
SORGAM,
SORASY etc which make
files to use as input to least
squares programs now automatically add these lines.
This information is used when necessary to calculate the parameters used in
Becker Coppens extinction so these no longer need be included with the data -
(L MODE=5 and L MODE=7 are now obsolete).
The data are now read only once, rather than once per least squares cycle as
in versions prior to 4.4.
The least squares programs
SFLSQ,
MPLSQ,
MAGLSQ,
MMPLSQ,
CHILSQ,
MPCLSQ and
MGTLSQ
all now use this new structure.
-
SFLSQ and
MPLSQ can only read structure factor type data (REFI 1-4)
-
CHILSQ and
MPCLSQ normally only read polarised neutron data (REFI=5 or 7)
but in both cases data measured with different orientations or wavelengths
can be refined together. It is sometimes useful to include unpolarised structure factor data
measured with an applied field as well.
Update 4.4.17 allows polarised neutron data from polycrystalline samples of anisotropic
paramagnets to be refined (MODE 11, REFI 9)
-
MAGLSQ,
MMPLSQ and
MGTLSQ all allow integrated intensity and polarised neutron data to be combined. (REFI=1-4,5,and 7)
When there are multiple data sets, each is presented in a separate file and the
values of MODE and REFI for each as well as its allocated weight may be given at the same time as the file-name.
In this case the request for the data file name has the form:
Name of data set, its MODE, REFI and weight? :
If the reply given is just the filename then the program assumes there is just a
single source of data: MODE and REFI are taken from the crystal data file
as in the pre-4.4 versions.
In the new program
SNPLSQ, which allows SNP and structure factor data to be refined
together, provision has to be made for different scale factors and different
magnetic domain populations to be associated with each data-set. These quantities must
also be identified with valid least squares parameters. Each data set is therefore identified by a
name of up to 4 characters eg PA27 which is given on a L SORC card. SORC
may be followed by the filename given with a path which is either absolute (starts with ``/")
or relative to the current working directory. Environment variables are recognised.
If no filename follows the identifier, the file name will be requested interactively.
For each data source identified by name there should be one or more L SORC name cards
giving further information specific to these data: REFI , MODE and WGHT are obligatory. The
integers following REFI and MODE have their usual meanings but apply only to this particular data-set. The number following WGHT gives the weight for these data.
There should also
be L SORC SCAL cards and L SORC DPOP cards giving the scale factors and domain populations
for the data set. The number of scale factors needed is the number of scaling zones
given in the data file. The number of domains whose populations must be given depends
on the relative symmetries of the nuclear and magnetic structures and on the type of refinement.
The new least squares parameters associated with the scales and domain populations are identified by the genus name and a composite species name starting with either SC for a scale factor
or DP for a domain population and ending with 2 digits identifying its number.
L DATA PA27 $TT/khe27k.pal
L SORC PA27 MODE 9 REFI 8 WGHT 1.0
L SORC PA27 DPOP 0.18 0.07 0.07 0.18 0.18 0.07 0.07 0.18
L DATA SF30 $TT/khe30k.sf
L SORC SF30 MODE 7 REFI 1 WGHT 0.3 SCAL 7.16
L SORC SF30 DPOP 0.25 0.25 0.25 0.25
The data are for a helical structure with 4 orientation domains. PA27 contains
SNP data (REFI 8) which is sensitive to the chirality so 8 domain populations are needed.
SF27 is a set of structure factor amplitudes (REFI 1) insensitive to chirality so only 4
domain populations can be refined. Pairs of chiral domains are numbered consecutively
so only odd-numbered domains are defined for the SF27 data. Constraints might therefore be
given as follows:
Z Domain constraints for PA data
L RELA 1 1 PA27 DP08 1 PA27 DP01
L RELA 1 1 PA27 DP07 1 PA27 DP02
L RELA 1 1 PA27 DP06 1 PA27 DP03
L RELA 1 1 PA27 DP05 1 PA27 DP04
L RELA 2 1 PA27 DP01 1 PA27 DP02 1 PA27 DP03 1 PA27 DP04
Z Domain constraints for SF data
L RELA 2 1 SF30 DP01 1 SF30 DP03 1 SF30 DP05 1 SF30 DP07
No scale factors are needed for the SNP data; one is needed for the structure amplitudes.
It could be fixed using:
L FIX SF30 SC01
For least squares refinements using different types of data, the weight given
to each is important. A rough rule of thumb is to make the relative weights
inversely proportional to the residual 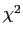 of each when refined separately.
of each when refined separately.



Next: MAGNETIC STRUCTURES AND STRUCTURE FACTORS
Up: LEAST SQUARES REFINEMENT USING
Previous: Making Other Least Squares
P.J. Brown - Institut Laue Langevin, Grenoble, FRANCE.
![]() of each when refined separately.
of each when refined separately.