D22
Large dynamic range small-angle diffractometer
Documentation

This is the home page of D22, the latest small-angle neutron scattering instrument at the ILL.
Inside the ILL, you can get the up-to-date version of this help file from the D22 server.
- Polarisation and polarisation analysis are available on D22. (18-Jun-2007)
- D22 has been equipped with additional security system during the 2004/2005 winter shutdown.
The interlock system now requires pushing a button in the controlled area after checking the absence of any person before opening the beam. (04-Mar-2005) - Instructions in case of problems with the detector high voltage (i.e. no detector image or no detector movement possible) and with missing parts of the detector image (27-Jul-2004)
- A new fast detector has been installed on D22 during the 2003/2004 winter shutdown.
It is made up of 128 linear sensitive Reuter-Stokes tubes of 8 mm diameter with 15 bars of 3He. Each tube can count neutrons at a rate of 50 kHz with 10% dead time losses.
(16-Mar-2004)
New on D22
Manuals | D22 information | General information |
|---|---|---|
| D22 publications | Instrument control electronics | |
| Important telephone numbers | Data analysis & treatment (Ron Ghosh) | |
| D22 computing environment | GRASP | |
D22 GEORGE dials | Pump control | |
Changer File, KIN, Q range etc. | Run parameters stored with the data | |
| Logicals used in movements on D22 | ||
| Interlock system | ||
| Contents of D22 cupboards | ||
| PF1 induced background | ||
| Fast Detector | ILL phone book (on site only) | |
D22's robot | ||
Web document produced by Roland May, ILL Grenoble (last update: 18-Jun-2007)
State of the Art
Roland P. May Institut Max von Laue - Paul Langevin, F-38042 Grenoble Cedex 9
1. Introduction
1.1. Small-angle (neutron) scattering
SANS is produced by heterogeneities in matter (that can be as well defined as identical viruses in aqueous solution). If these are randomly oriented, every atom pair contributes a sin (Qd) / (Qd) term to the scattering of a sample, where d is the distance between the two atoms of the pair, and Q the momentum transfer, Q = (4π/λ) sin Θ, with 2Θ, the full scattering angle and λ, the wavelength.
Inhomogeneities of sizes larger than atomic distances (10 to 1000 Å) produce scattering patterns with interesting Q ranges 1/D < Q < 10/D, if D is the dimension of the inhomogeneities. The scattering angle 2Θ corresponding to the upper Q limit for D=100 Å is about 9.1 ° for neutrons of 10 Å wavelength.
In most cases, the sample and/or the sample environment are rather bulky. Therefore, SANS instruments usually have to be large themselves in order to yield the desired resolution. Small SANS instruments can only serve a very limited number of applications. For reasons of intensity, a relatively large beam divergence, i.e. a beam cross-section larger than the sample size is accepted as well as wavelength resolutions Δλ/λ of up to about 20 %.
1.2. Neutron properties
Discovered by Chadwick in 1932
Rest mass | Temperature dependence of the wavelength |
Electric charge | Life time |
Magnetic moment | Frequency |
Speed | Spin 1/2 |
Total reflection: Ultra-cold neutrons, neutron guides
2. History
1983: Proposal (R.P. May, R. Oberthür, P.A. Timmins and A.F.Wright)
1987 (September): Final proposal (R.P. May, M. Thomas) endorsed by Instrument Subcommittee
1989 (December): Delivery of detector tube and collimation by ABT (SIGRI)
1992 (December): Delivery of detector by CERCA (after 24 months)
1995 (February): First tests after reactor overhaul: first users in April
1996 (April): Start of permanent operation after repair of detector
3. Location
3.1. Guide Hall
3.2. View of D22

4. Features
4.1. Wavelength selection and collimation
"Classical" pin-hole small-angle scattering instrument
Wavelength selection
- DORNIER velocity selector 25 cm long, 28 300 rpm, resolution 10% (fwhm), 4.6 < l < 42 Å for 0 ° tilt
Collimation system
- guide cross-section 55 x 40 mm
- 8 drums of lengths varying as a geometical series to yield free flight paths of 1.4 to 17.6 m
- three positions: guide in / apertures (/ optical elements)
- 3 attenuators and 4 round apertures (5, 10, 20, 30 mm diameter), computer controlled
- manually remotely controlled aperture at 19.1 m from sample
4.2. Detector
- 3He, 2 bars; sensitive surface: 0.96 m x 0.96 m
- 16 K pixels of 0.75 x 0.75 cm
- minimal distance 1.35 m, maximal distance 18 m
- can be translated by half its width, i.e. 50 cm
- can be rotated around its vertical axis (25 °)
- Q-range: 0.7x10-3 Å-1< Q < 1 Å -1
Qmax/Qmin up to 50, typically 20 with offset, 10 without
4.3. Sample position
- table for mounting equipment in air, e.g. sample changer, shear apparatus
- vessel ("cloche") for working in vacuum
- electromagnets (1.4 T)
- furnace (up to 1300 °C)
- "orange" cryostats with support permitting direct-beam and reference measurements
- remotely controlled heating/cooling bath (about -10 / +80 °C)
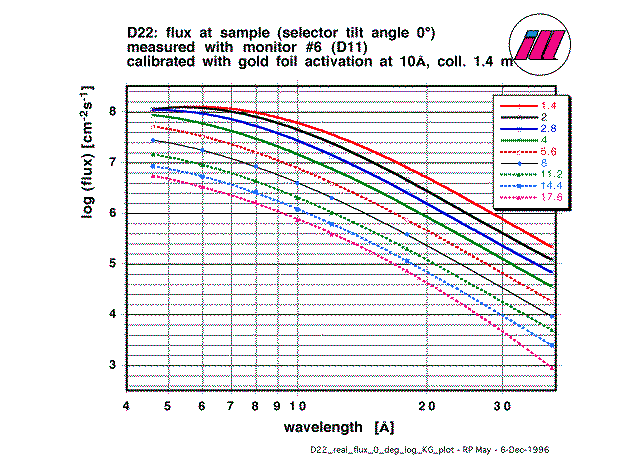
4.4. Flux
4.5. Sample zone and sample-preparation lab


4.6. Cryostats and rack plate



4.7. Robot
A robot arm for manipulating radioactive (and other) samples


5. Experiments
5.1. Proliferating Cell Nuclear Antigen (PCNA)
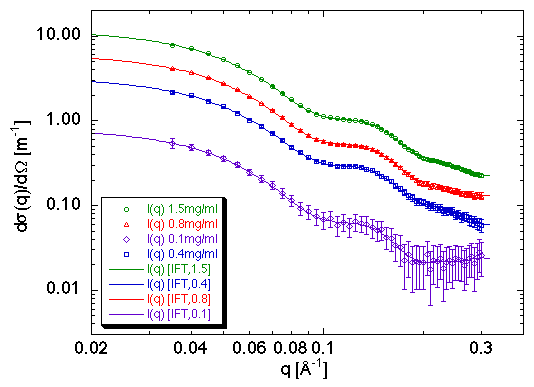
P. Schurtenberger et al. (1998) J. Mol. Biol. 275, 123-132
5.2. The diffusion of oxygen in silicon single-crystals

S. Rycroft and R.J. Stewart (Univ. Reading)
The small-angle neutron-scattering from SiO2 precipitates in a single crystal of silicon which has been heat treated for 500 hours at 600°C; the neutron beam was incident along a <100> direction. The central cross arises from the cushion shaped SiO2 precipitates lying on (100) planes with their edges along <110> directions.
5.3. Nucleation and growth studied by time-resolved SANS
Homogeneous nucleation and growth of oil droplets studied by time-resolved small-angle neutron-scattering.
S.U. Egelhaaf (ILL), P. Schurtenberger (ETH Zürich), J. Morris, U. Olsson, H. Wennerström (Lund University)
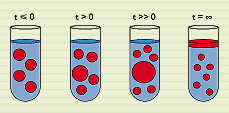
Figure 1
Oligodisperse oil-in- water microemulsion droplets are quenched into a two phase area. At final equi- librium, smaller droplets coexist with an excess oil phase that nucleates at a few of the initial droplets which subsequently grow (t > 0), allowing the majority of droplets to decrease in size. This growth phase proceeds with a constant number of large drops (t >> 0).
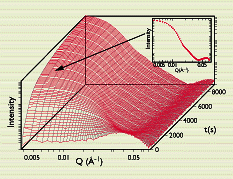
Figure 2
Sequence of scattering curves from a time-resolved small-angle neutron-scattering experiment recorded during the nucleation and growth of an excess oil phase from oil-in-water microemulsion droplets. For clarity only every second measurement is shown. As an example the scattering curve obtained at t = 3150 s is added as an inset.
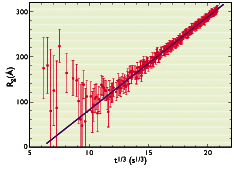
Figure 3
Time-dependence of the radius of gyration Rg,big as obtained from a Guinier fit. Also shown is the power law behaviour expected for a classical Ostwald ripening process.
5.4. The binding of GroEL and GroES (time-resolved SANS)
Jörg Holzinger and Roland May (ILL), Manfred Rößle, Elena Manakova and Hermann Heumann (MPI für Biochemie)
5.5. He bubble growth in a fusion-reactor steel
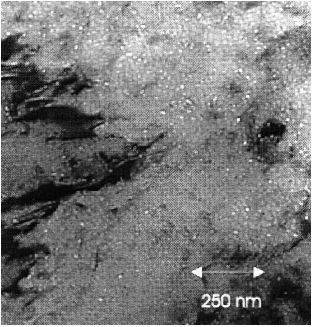
Figure 1
Transmission electron micrograph of He bubbles after implantation of 400 appm He at 250 °C (from the work of Bertsch, 1997).

Figure 2
Nuclear plus magnetic cross sections of reference and implanted samples for 250 °C (full circles), 825 °C (empty circles), and 975 °C (squares).
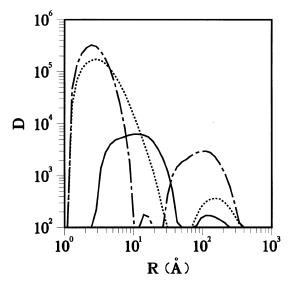
Figure 3
He-bubble volume distributions (a.u. proportional to bubble volume per unit volume vs. bubble size in Å) obtained from the data of Fig. 2: 250 °C (dotted), 825 ° C (dash-dot), and 975 °C (solid).
Helium Bubble Growth in a Martensitic Steel for Fusion Reactors
6. Projects
Real-time SANS
- The high flux allows us to perform real-time experiments
- A much faster detector is needed to work correctly at large Q
Polarization & polarization analysis
- Polarized neutrons were used by Neumann et al.
- Werner Heil is interested in developing a 3He spin filter
Spin contrast variation
- Oliver Zimmer is adapting a polarized target station
- A test experiment to look at the sizes and the time behaviour of polarized regions in polymers and proteins is planned
Time-of-flight SANS
- Space for three chopper systems exists in the design of D22
7. People
Design | Detector | Electronics | Hall d'essais | Mechanics | Neutron distribution | Organisation | Programming | Technicians |
|---|---|---|---|---|---|---|---|---|
Klaus Gobrecht | Roger Chung Michel Gamon Bruno Guérard Jean Jacobé Anton Oed André Rambaud Jochen Uckelmann | Antony Burton Frédéric Descamps Reinhard Klesse Jacques Munnier Jacques Ratel Franck Rey J.-Antoine Vidal-Garcia | Claude Gomez Marc Locatelli Anthony Pascaud Émile Stropiano Pierre Thomas | Tony Billington J.-François Carliez Luc Didier Guy Gobert Michel Goursaud Erwin Hetzler Jacques Loppé Philippe Malbert | Alain Beynet Peter Suttling Jean-Rémy Villard | Jean-Claude Faudou Marie-Claude Filhol Bernard Guérra Walther Kaiser | Alain Barthélémy Franck Cécillon Ron Ghosh Georges Messoumian Didier Richard Michel Roure Helga Schwab Odile Tillier Klaus Wotschack | Michel Bonnaud |
and many others...
Fast Detector
Welcome on the D22 Millenium Project homepage
Supported by TECHNI (Technology for Neutron Instrumentation)
Project Plan in its initial version (April 2000)
The aim of the project is to develop and to build a real-time neutron detector for small-angle scattering applications which is capable of counting 2 MHz of neutrons on the whole detector with dead-time losses of not more than 10%, rather than the 50 KHz of the current detector. In all other aspects the new detector should be at least as efficient as the current detector of the instrument D22 at the ILL, i.e. it should in particular have a spatial resolution of 7.5 x 7.5 mm, an active area of 96 x96 cm, a sensitivity of 75% for neutrons of 5Å wavelength, and a similar quiet count rate.
We envisage two possible solutions to achieve this goal. The first is that of an "organ-pipe" detector composed of about 120 single linear position sensitive detectors arranged in a vertical plane or (more probably) in a zig-zag. The second is an array of linear position sensitive wires situated in a housing similar to the current detector. These wires would have to be on an insulated "comb" that replaces the current anode plane. The latter solution requires development of electronics to determinate the lateral centre of gravity of detection events.
The project is split in two phases, an exploratory phase of two years to select the better solution and to test a prototype, and a construction phase for producing such a detector for D22. A similar but smaller detector for D11 would follow as a separate project. The cost of phase I, wich is to be completed by December 2001, is estimated as 560 kF plus 76 weeks of manpower, that phase II, to be completed by December 2003 or June 2004, to 3640 kF plus 30 weeks of manpower.
Risks: after two years R&D the required 2 MHz count rate may not be achievable whilst retaining all other characteristics of the present detector. At this point, a decision would be made on whether to proceed with a reduced specification of the detector.
Project status in November 2003
The solution of independant PSDs has been rapidly prefered to the Multi-wire single vessel detector for reasons of simplicity and cost. In the zig-zag configuration, the shadowing effect of detectors located in the front plan requires a calibration which depends on the distance between the sample and the detector, and of the neutron wavelength. For this reason, the zig-zag configuration, which was first considered to maximise detection efficiency, will not be implemented . Several prototypes of the D22 PSD have been produced by Reuter Stokes, and extensively studied at the ILL. Specifications are now freezed.
Detection performances are matching the D22 specifications:
70% efficiency at 6 Angstroms (taking into account the dead zones between tubes)
6 mm FWHM position resolution along the tubes
100 KHz/PSD counting rate.
The mounting of the detector mechanics will start in December 2003, and we plan to start the commissioning phase at the begining of the next reactor cycle, in March 2004.
GRASP
SAXS/SANS Platform
Welcome to the Small Angle Scattering Platform for Structural Biology!
Small-Angle Scattering is a powerful technique to study macromolecule conformations in solution. X-rays and Neutrons scattering present very complementary aspects. In order to learn how these techniques can enrich your research project, what are their specificities and how to prepare a SAXS or SANS experiment, please read the instructions contained in this page. Local scientists in charge of BioSAXS (BM29 at ESRF) and BioSANS (D22 at ILL) instruments would be pleased to help you if you need further details or if you plan to submit an experiment proposal.
Be aware that in case you would like to take advantage of the complementarity of these two methods, it is possible to submit a joint SAXS-SANS proposal: you should write a SANS proposal in which you precise that you also want to caracterise your sample by SAXS.
To contact responsible scientists:
BioSANS:
Anne Martel
Frank Gabel
BioSAXS:
Petra Pernot
Martha Brennich
Introduction to Small Angle Scattering (SAS) for Biological Macromolecular Complexes
In the last decade, the number of high resolution protein structures has increased drastically as seen from the Protein Data Bank which contains now more than 30 000 structures. Tremendous progress in the automation of protein overproduction, crystallization, data collection and data treatment has facilitated the crystallographic studies of macromolecules. The challenge for the next decade is to integrate the molecular descriptions with their cellular environment. For this, structures of larger macromolecular complexes have to be deciphered. These multi-component complexes often participate in signal transduction pathways and have therefore dynamic structures that have to be unravelled by multiple approaches combining various resolutions and spatial scales.
Crystallographic approaches are very powerful in solving structures of proteins or macromolecular complexes at atomic resolution when they adopt well-defined conformations and can therefore be crystallized.More labile conformations, characteristic of modular proteins or transient complexes have to be characterized by other approaches. The combination of high-resolution techniques with low or medium resolution approaches leads to quasi-atomic models. This is well illustrated by several examples of virus structures where electron microscopy on single viruses provides for the overall viral envelope and X-ray crystallography of individual proteins leads to atomic structures that can be fitted into these envelopes. Small angle scattering has been used since several decades, although its use was very often limited to very low resolution, the so-called Guinier region, analysis of which allows the determination of the radius of gyration and the molecular mass. In the last few years, experimental X-ray set-ups have improved and higher resolution scattering data are now exploitable.
Further mathematical developments made it possible to fit a molecular envelope to the interatomic distance distribution, the P(r) function, deduced from the small angle scattering data1. Small angle X-ray scattering is now a powerful tool for determining molecular envelopes that can be combined with high-resolution structures. In contrast to electron microscopy, the method can be used for smaller macromolecules and is therefore complementary. Neutron small angle scattering provides additional information for macromolecular complexes that are made of several types of molecules such as proteins, nucleic acids or lipids. Contrast variation experiments obtained by exchanging the solvent for deuterated or partially deuterated solvent enhances the signal from one component. SAXS and SANS are therefore tools of major importance in tackling systems biology.
Neutron small angle scattering instruments have been developed at ILL since several years and are available to the community. An X-ray small angle instrument is partially available for biological studies at ESRF, but does not respond to the increasing demand of the scientific community. This project aims to reinforce the development of small angle scattering for biological studies in general in Grenoble, by improving the instrumental facilities and helping in good quality data collection. It should also help in bridging the gap between the experimental data and the final molecular modeling in combination with X-ray or NMR partial structures.
References
1. Petoukhov, M.V. & Svergun, D.I. (2005)
Global rigid body modeling of macromolecular complexes against small-angle scattering data.
Biophys J 89(2):1237-50.
2.Svergun, D.I. & Koch, M.H.
Advances in structure analysis using small-angle scattering in solution.
Curr Opin Struct Biol 2002 12(5):654-60.
3.Grizot S, Grandvaux N, Fieschi F, Faure J, Massenet C, Andrieu JP, Fuchs A, Vignais PV, Timmins PA, Dagher, MC, Pebay-Peyroula E. (2001)
Small angle neutron scattering and gel filtration analyses of neutrophil NADPH oxidase cytosolic factors highlight the role of the C-terminal end of p47(phox) in the association with p40(phox).
Biochemistry. 40, 3127-33
4. Durand, D., Cannella, D., Dubosclard, V., Pebay-Peyroula, E., Vachette, P., and Fieschi, F. (2006).
Small-Angle X-Ray Scattering Study reveals an Extended Organization For The Auto-Inhibitory Resting State of the p47phox Modular Protein.
Biochemistry 45, 7185-7193
5. Gutsche I., Holzinger J., Rauh N., Baumeister W., May R.P. (2001)
ATP-Induced Structural Change of the Thermosome Is Temperature-Dependent
Journal of Structural Biology, 135, Number 2, 139-146(8)
6. King W.A., Stone D.B., Timmins P.A., Narayanan T., von Brasch A.A.M., Mendelson R.A., Curmi P.M.G. (2005)
Solution structure of the chicken skeletal muscle troponin complex via small-angle neutron and X-ray scattering
J. Mol. Biol. 345, 797-815
7. Heller, W.T., Finley, N, Wen-Ji D., Timmins, P., Cheung, H.C., Rosevear, P.R. and Trewhella, J (2003)
Small-angle neutron scattering with contrast variation reveals spatial relationships between the three subunits in the ternary troponin complex and the effects of troponin I phosphorylation
Biochemistry 42(25):7790-7800
8. Fukuhara, N. Conti, E., Svergun, D., Petoukhov, P. and Timmins, P. (in preparation)
Structure of the tRNA nuclear export complex
9. Callow P., Timmins P. and Kneale G. (2006)
Preliminary neutron scattering studies of the Type I restriction-modification enzyme M.Ahdl
Physica B - Vol.385-386 - pp.853-855
by P. Timmins (ILL)
Practical guidelines for SAXS/SANS users
1) Getting beamtime (proposals, reports and deadlines): Access to beamtime at ESRF (http://www.esrf.eu/UsersAndScience/UserGuide/Applying) and ILL (http://www.ill.eu/users/applying-for-beamtime/electronic-proposal-system/) is granted via electronic proposal systems. Potential users requesting beamtime must submit a scientific proposal online, providing scientific background on the experiments, description of the samples to be measured, time requested etc. The systems are generally open twice a year with deadlines that must be respected (at ESRF, there is a more frequent request of beamtime possible, the so-called rolling beamtime (http://www.esrf.eu/UsersAndScience/UserGuide/Applying/ProposalGuidelines/MXnon-BAGproposal/macromolecular-crystallography-rolling-applications)). In a proposal, a local contact that accompanies you during the experiment must be indicated in general. If successful, beamtime is generally attributed during the 6 months that follow the proposal deadline. The submitted proposals are reviewed by scientific committees that evaluate their scientific quality. Due to an overload factor that varies from instrument to instrument, not all proposals can be accepted. If beamtime has been granted, the users will be notified. Additional information about travel, accommodation and reimbursement can be found at the ILL (http://www.ill.eu/users/user-guide/) and ESRF (http://www.esrf.eu/UsersAndScience/UserGuide).
Joint SANS/SAXS proposals are possible; the request for additional SAXS beamtime must be indicated in your the ILL SANS proposal submission. If the SANS part of the proposal is accepted by the ILL subcommittee, you need to submit a request for the SAXS beamtime at ESRF (via the rolling beamtime system).
2) Before the experiment (sample preparation and pre-characterization): The key to a successful SAXS/SANS experiment in biology is to know the biochemistry of the system studied. Prior to a SAXS/SANS experiment, as much information as possible should be collected on the system investigated by complementary methods. In the case of shape reconstruction, form factor analysis etc, monodisperse samples are needed (e.g. only one type of particle in solution). This should be checked by gel filtration, light scattering, SEC-MALLS, analytical ultracentrifugation, etc… The Partnership for Structural Biology (PSB) and the Partnership for Soft Condensed Matter (PSCM) offer a large variety of platforms for sample characterization (http://www.psb-grenoble.eu/spip.php?rubrique80 ; http://www.ill.eu/instruments-support/labs-facilities/pscm/soft-matter-laboratory/). If you need access to them, please do contact the platform responsibles well in advance of your scheduled SAXS/SANS experiments.
The following points are particularly important:
- Aggregation state: structural studies on aggregated samples are in general not possible, it is therefore essential to avoid aggregation. SANS practitioners should be aware that an elevated D2O content in the solution (as required for some experiments) may favor aggregation. In general, higher sample concentrations also favor aggregation. In such cases it is advisable to run a concentration series starting at higher concentrations (~10mg/ml) and going down to smaller concentrations (~1mg/ml). In the end, the highest concentrated sample that shows no aggregation is evaluated.
- Concentration of the samples: concentrations for diluted samples measured by SAXS/SANS are typically situated in the range from 1-10 mg/ml (this corresponds to molarities of 0.05-0.5 mM for a 20 kDa system and 0.01-0.1 mM for a 100 kDa system). An accurate determination of the concentration prior to the experiments is also important for an absolute measurement of the molecular masses in solution by SANS. Concentrations should be known at an accuracy of about +/- 10%. In general, optical density measurements (e.g. at 280 nm for proteins) are more accurate than other assays (Bradford etc…).
- Sample volumes required: Samples at the indicated concentrations are needed in ~200 µl volumes for SANS (generally measured in quartz cuvettes) and in smaller amounts (~50 µl ) for SAXS (capillary injection system).
- Buffers: Buffers identical to the ones used with the samples (regarding salinity, pH, D2O content etc) need to be measured in order to subtract the background from the sample runs and to focus on the signal by the macromolecules in solution.
- Concluding remarks regarding samples: For more detailed information, see Jeffries et al. Nature Protocols 11, 2122–2153 (2016). It is generally advisable to bring some backup samples to a SAXS/SANS experiment in the case that some samples do not behave properly (aggregation etc…) and to discuss sample conditions with your local contact.
3) During the experiment: Depending on the size of the macromolecules studied, the sample volume available, the temperature and pressure required, the instrument needs to be set up specifically.
- Angular range (SANS): Collimator-sample and detector-sample distances need to be optimized against the size (i.e. presumed radius of gyration) of the macromolecules under investigation. If large objects are studied, these distances will generally be chosen large, if smaller objects and higher angular ranges are required, these distances will be chosen shorter.
- Instrument calibration: Before measuring samples, the small angle scattering instrument needs to be calibrated at a given detector-collimator setup: centering of the incoming neutron/X-ray beam, determination of the detector efficiency, evaluation of the background noise and determination of the signal from the sample holder / capillary device.
- Output of an experiment: The raw data from an experiment of an isotropically scattering system represent the scattering of the macromolecules in solution. They are generally corrected for the sample and instrument geometry, detector efficiency, electronic and buffer backgrounds, and averaged isotropically at a given angle. These operations are done by program suites that are specific for individual instruments.
4) After the experiment (sophisticated data treatment and interpretation):
- Integral parameters: Integral parameters such as radii of gyration and molecular weights of the objects studied are available directly from the data as described above.
- Modeling: The reduced raw data may be used to extract information going beyond radii of gyration and molecular weights. Several program suites are available to perform more sophisticated operations on the data (e.g. http://www.embl-hamburg.de/biosaxs/software.html). These include: extraction of a pair distribution function, ab intio low resolution modeling, rigid body modeling, etc.
Publications
Pérard J, Leyrat C, Baudin F, Drouet E, Jamin M. Structure of the full-length HCV IRES in solution.
Nat Commun. 2013 Mar 19;4:1612. doi: 10.1038/ncomms2611. pdf (pdf - 1.62 Mi)
F. Spinozzi , P. Mariani, I. Mičetić, C. Ferrero, D. Pontoni, M. Beltramini, " Quaternary structure heterogeneity of oligomeric proteins: a SAXS and SANS study of the dissociation products of Octopus vulgaris hemocyanin”, PLoS One 11, e49644 (2012). pdf (pdf - 1.11 Mi)
F. Spinozzi (a), P. Mariani (a), I. Mičetić (b,c), C. Ferrero (d), D. Pontoni (d), M. Beltramini (b),
Blue blood: a challenge for structural biologists. ESRF spotlight175 (2012). pdf (pdf - 144 Ki)
Leyrat C, Schneider R, Ribeiro EA Jr, Yabukarski F, Yao M, Gérard FC, Jensen MR, Ruigrok RW, Blackledge M, Jamin M.
Ensemble structure of the modular and flexible full-length vesicular stomatitis virus phosphoprotein.
J Mol Biol. 2012 Oct 19;423(2):182-97. pdf (pdf - 1.32 Mi)
E. Baldassarri Jr., M.G. Ortore, C. Ferrero, S. Finet, F. Spinozzi, P. Mariani, “Pressure effects on G-quadruplex structures stabilized by two different counter-ions”, International Review of Biophysical Chemistry (IREBIC), Vol. 2, N. 4, Special Section on 3rd International Meeting on G-Quadruplexes and G-assembly, p. 142, August 2011.
